Home
The Kuhlman Lab
Currently we are focusing on a variety of design goals including the creation of novel protein-protein interactions, protein structures and light activatable protein switches. Central to all of our projects is the Rosetta program for protein modeling. In collaboration with developers from a variety of universities, we are continually adding new features to Rosetta as well as testing it on new problems.
Recent Highlights
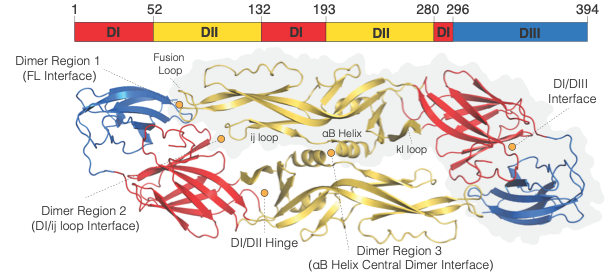
Dengue Engineering
Science Advances, 2021 Designed, highly expressing, thermostable dengue virus 2 envelope protein dimers elicit quaternary epitope antibodies Stephan Kudlacek, Stefan Metz, …. , Aravinda de Silva, Brian Kuhlman Dengue virus (DENV) is a worldwide health burden, and a safe … Read more
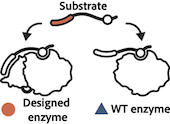
Enzyme Turbocharging
Nature Chemical Biology, 2023, 19(4):460-467 Designer installation of a substrate recruitment domain to tailor enzyme specificity. Park R, Ongpipattanakul C, Nair SK, Bowers AA, Kuhlman B. Promiscuous enzymes that modify peptides and proteins are powerful tools for labeling biomolecules; … Read more
Light Controllable Nuclear Export
Nat Chem Biol. 2016 Apr 18 Light-induced nuclear export reveals rapid dynamics of epigenetic modifications. Yumerefendi H, Lerner AM, Zimmerman SP, Hahn K, Bear JE, Strahl BD, Kuhlman B. We engineered a photoactivatable system for rapidly and reversibly exporting proteins from … Read more
Protein SEWING
Science. 2016;352(6286):687-90 Design of structurally distinct proteins using strategies inspired by evolution. Jacobs TM, Williams B, Williams T, Xu X, Eletsky A, Federizon JF, Szyperski T, Kuhlman B. Natural recombination combines pieces of preexisting proteins to create new tertiary structures and … Read more